EMR Series 2: Running and troubleshooting jobs in an EMR cluster
This is the second installment of our three-part blog series on how to send Amazon Elastic MapReduce (EMR) log files to SolarWinds® Loggly®. In the first part, we provided a quick introduction to EMR, Amazon’s distribution of Apache Hadoop. An EMR cluster is a managed environment that differs slightly from a traditional Hadoop cluster. We saw how EMR generates different types of log files and where those logs are saved. These log files are invaluable when troubleshooting failed Hadoop jobs
In this post, we will create an EMR cluster and configure it to run multi-step Apache Hive jobs. We will intentionally introduce an error in one of the steps, and use EMR log files to find the root cause
The Scenario
The EMR cluster will have Apache Hive installed in it. This cluster will use EMRFS as the file system, so its data input and output locations will be mapped to an S3 bucket. The cluster will also use the same S3 bucket for storing log files
We will create a number of EMR steps in the cluster to process a sample set of data. Each of these steps will run a Hive script, and the final output will be saved to the S3 bucket. These steps will generate MapReduce logs because Hive commands are translated to MapReduce jobs at run time. The log files for each step will be aggregated from the containers it spawns
Sample Data
The sample data set for this use case is publicly available from the Australian government’s open data website. This data set is about threatened animal and plant species from different states and territories in Australia. A description about the fields of this data set and the CSV file can be seen and downloaded here.
Processing Steps
The first EMR job step involves creating a Hive table as a schema for the underlying source file in S3. In the second job step we will run a successful query against the data. We will then run a third and fourth query; the third one will fail and the fourth will succeed
We will repeat these four steps a few times in an hour, simulating successive runs of a multi-step batch job. However, in a real-life scenario, the time difference between each batch run could be much higher. The small time gap between successive runs here is intended to expedite our testing
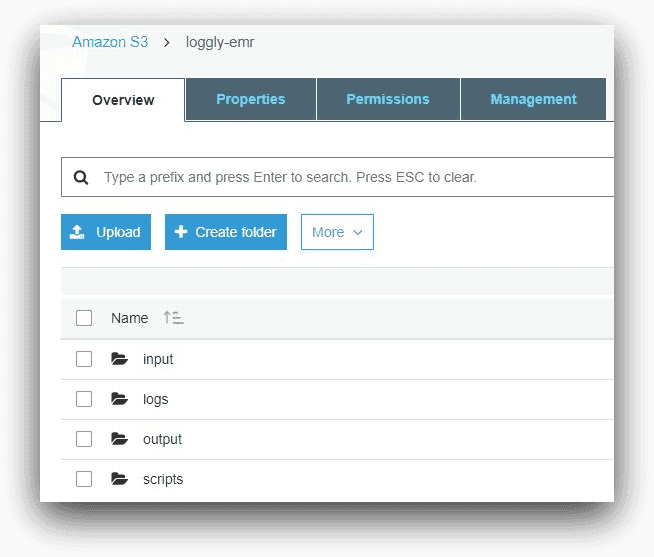
S3 Bucket and Folders
Before creating our EMR cluster, we had to create an S3 bucket to host its files. In our example, we have named this bucket “loggly-emr.” The folders under this bucket are shown below in the AWS Console for S3:
- The input folder holds the sample data
- The scripts folder contains the Hive script files for EMR job steps
- The output folder will hold the Hive program output
- The logs folder will be used by the EMR cluster to save its log files. As we saw in the first part of this series, EMR will create a number of sub-folders inside this folder. The logs from the logs folder will be ingested into Loggly.
Hive Scripts for EMR Job Steps
Step 1
This job step runs a Hive script createTable.q to create an external Hive table. This table describes the tabular schema of the underlying CSV data file. The script is shown below:
CREATE EXTERNAL TABLE `threatened_species`( `scientific name` string, `common name` string, `current scientific name` string, `threatened status` string, `act` string, `nsw` string, `nt` string, `qld` string, `sa` string, `tas` string, `vic` string, `wa` string, `aci` string, `cki` string, `ci` string, `csi` string, `jbt` string, `nfi` string, `hmi` string, `aat` string, `cma` string, `listed sprat taxonid` bigint, `current sprat taxonid` bigint, `kingdom` string, `class` string, `profile` string, `date extracted` string, `nsl name` string, `family` string, `genus` string, `species` string, `infraspecific rank` string, `infraspecies` string, `species author` string, `infraspecies author` string) ROW FORMAT DELIMITED FIELDS TERMINATED BY ',' STORED AS INPUTFORMAT 'org.apache.hadoop.mapred.TextInputFormat' OUTPUTFORMAT 'org.apache.hadoop.hive.ql.io.HiveIgnoreKeyTextOutputFormat' LOCATION 's3://loggly-emr/input/' |
Step 2
This job step runs a query to calculate the top five endangered species in the state of New South Wales (NSW). The Hive query file name is endangeredSpeciesNSW.q and it’s shown below:
SELECT species, COUNT(nsw)AS number_of_endangered_species FROM threatened_species WHERE (nsw = 'Yes' OR nsw = 'Endangered') AND "threatened status" = 'Endangered' GROUP BY species HAVING COUNT(nsw) > 1 ORDER BY number_of_endangered_species DESC LIMIT 5 |
Step 3
This job step runs a query to calculate the total number of endangered plant species for each plant family in Australia. However, this query has a deliberate error in the code where a field is not added to the GROUP BY clause. This is the error we want Loggly to highlight. This query will fail when Hive tries to run it. The Hive query file name is endangeredPlantSpecies.q and is shown below
SELECT family, COUNT(species) AS number_of_endangered_species FROM threatened_species2 WHERE kingdom = 'Plantae' AND "threatened status" = 'Endangered' |
Step 4
This step will again succeed. It lists the scientific names of extinct animal species in Australia’s Queensland state. The script file is called extinctAnimalsQLD.q and is shown below:
SELECT "common name", "scientific name" FROM threatened_species WHERE kingdom = 'Animalia' AND (qld = 'Yes' OR qld = 'Extinct') AND "threatened status" = 'Extinct' |
Log Aggregation
We have also uploaded a JSON file called logAggregation.json in the scripts folder of the S3 bucket. This file will be used for aggregating the YARN log files. Log aggregation is configured in in the yarn-site.xml configuration file when the cluster starts up. The contents of logAggregation.json file is shown below:
[
{
"Classification": "yarn-site",
"Properties": {
"yarn.log-aggregation-enable": "true",
"yarn.log-aggregation.retain-seconds": "-1",
"yarn.nodemanager.remote-app-log-dir": "s3:\/\/loggly-emr\/logs"
}
}
]
|
EMR Cluster Setup
After the S3 bucket is created and the data and script files are copied into their respective folders we set up an EMR cluster. The following images describe the process as we create the cluster with mostly default settings
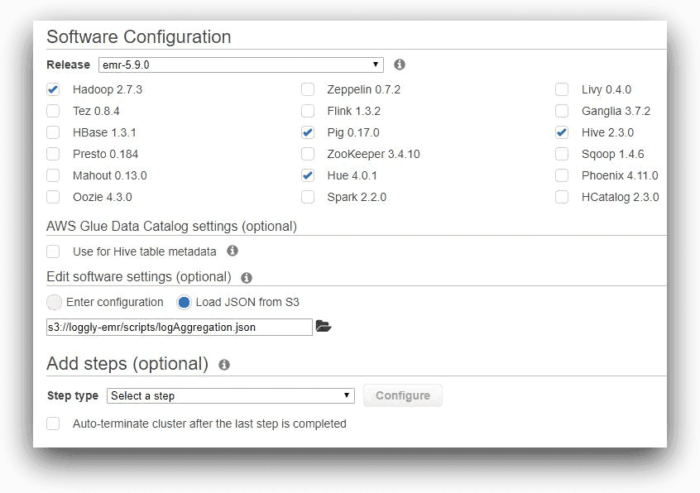
In the first screen, to configure the cluster in the AWS console, we have kept all of the applications recommended by EMR, including Hive. We are not using AWS Glue for storing Hive meta data, nor are we adding any job step at this time. However, we are adding a software setting for Hive. Note how we are specifying the path to the log aggregation JSON file in this field
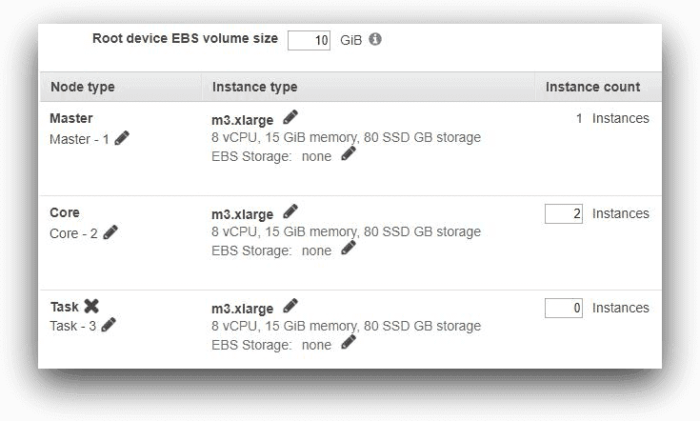
In the next screen, we have kept all the default settings. For the purpose of our test, the cluster will have one master node and two core nodes. Each node is an m3.xlarge instance and has 10 GB root volume.
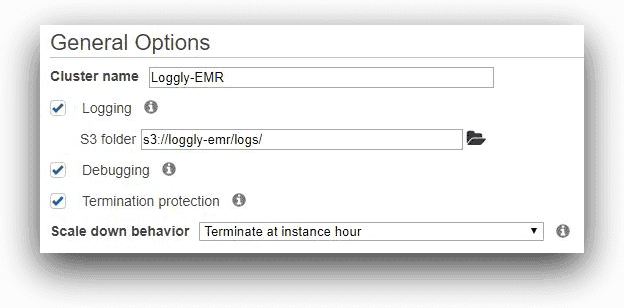
We are naming the cluster Loggly-EMR in the next screen, and specifying the custom s3 location for its log files
We are leaving default settings for EMRFS consistent view, custom AMI ID and bootstrap actions
Finally, we specified an EC2 key pair for accessing the cluster’s master node. The default IAM roles for EMR, EC2 instance profile, and auto-scale options are not changed. Also, the master and core nodes are using default security groups
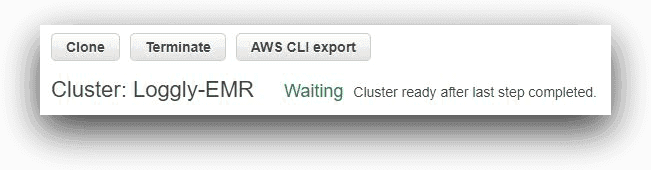
Overall, this is a default setup for an EMR cluster. Once ready, the cluster is in a “waiting” status as shown below:
Submit Hive Job Steps
With the EMR cluster up and running, we added four job steps from the command line using the AWS CLI’s aws emr add-steps command. Note how the script specifies the Hive script file and input and output folder for each step:
aws emr add-steps --cluster-id j-2TFSCG8AY15CK --steps Type=HIVE, Name='createTable', ActionOnFailure=CONTINUE, Args=[-f,s3://loggly-emr/scripts/createTable.q,-d,INPUT=s3://loggly-emr/input,-d,OUTPUT=s3://loggly-emr/output] Type=HIVE, Name='endangeredSpeciesNSW', ActionOnFailure=CONTINUE, Args=[-f,s3://loggly-emr/scripts/endangeredSpeciesNSW.q,-d,INPUT=s3://loggly-emr/input,-d,OUTPUT=s3://loggly-emr/output] Type=HIVE, Name='endangeredPlantSpecies', ActionOnFailure=CONTINUE, Args=[-f,s3://loggly-emr/scripts/endangeredPlantSpecies.q,-d,INPUT=s3://loggly-emr/input,-d,OUTPUT=s3://loggly-emr/output] Type=HIVE, Name='extinctAnimalsQLD', ActionOnFailure=CONTINUE, Args=[-f,s3://loggly-emr/scripts/extinctAnimalsQLD.q,-d,INPUT=s3://loggly-emr/input,-d,OUTPUT=s3://loggly-emr/output] |
There should be no line breaks when running this script. We have added the line breaks for clarity’s sake. The output of this command will be a list of job step IDs:
{
"StepIds": [
"s-27S2V7H36F11A",
"s-20S96C57D979O",
"s-1OZI9O3LPFMIH",
"s-2128EHDHIPD3M"
]
}
|
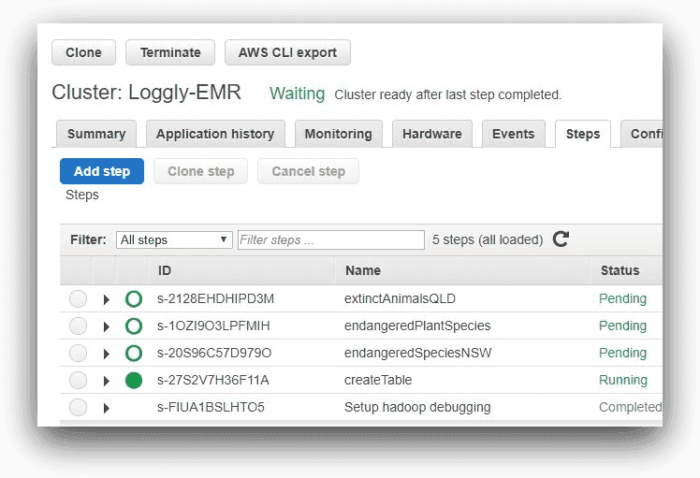
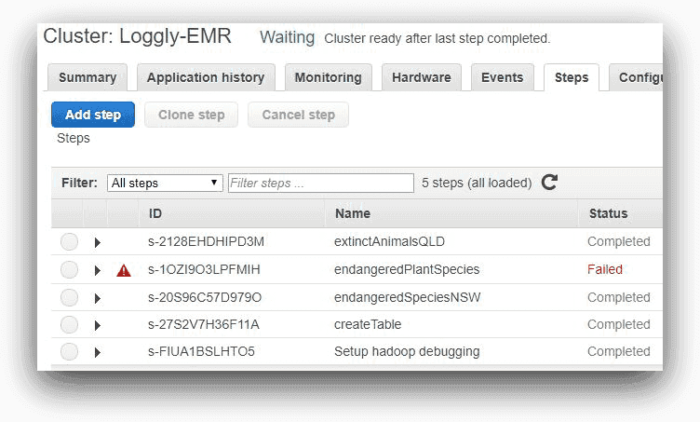
These are the steps EMR would run one after another. The image below shows the steps from AWS EMR console:
Here, one of the steps is running and the other three are pending. As each step completes, it will be marked as “Completed.”
As expected, the third will fail, as shown in the next screenshot
The log files for each step will be saved under subfolders in s3://loggly-emr/logs/<EMR-cluster-id>/steps folder
Each node will have its logs saved under s3://loggly-emr/logs/<EMR-cluster-id>/node folder
This image of the console reflects the status of each step after run:
Troubleshooting an Error Using the Log Files
Before using Loggly for any troubleshooting, let’s view these logs from the EMR console. We could also download the log files from the S3 folder and then open them in a text editor. For each step, the controller log file will contain the most relevant information
In the Steps tab of the cluster, if we clicked on the “controller” link for the failed job step, the log file will open in a separate browser tab
Two lines from this log file will be important. At the very beginning, the log will show a message like this:
INFO startExec 'hadoop jar /var/lib/aws/emr/step-runner/hadoop-jars/command-runner.jar hive-script --run-hive-script --args -f s3://loggly-emr/scripts/endangeredPlantSpecies.q -d INPUT=s3://loggly-emr/input -d OUTPUT=s3://loggly-emr/output' |
This shows the name of the Hive script that the Hadoop command runner is starting.
Near the end of the file, there will be another message like this:
WARN Step failed with exitCode <nn> |
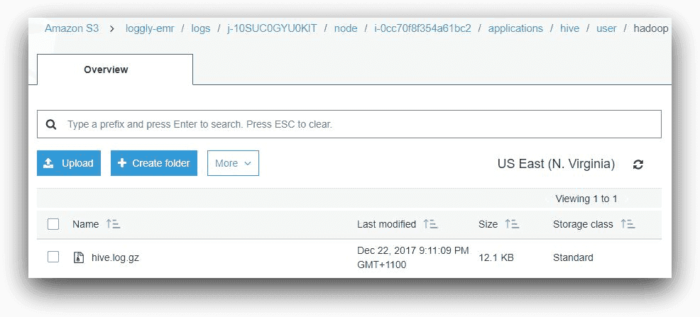
This shows that the step has failed. But why has it failed? To find the answer, we can look under the Hive application subfolder in the logs folder. This folder will contain a file called hive.log.gz. The Hive log file contains detailed information about each HiveQL command executed. In our case, the file is in the following location:
s3://loggly-emr/logs/<cluster-id>/node/<master-node-id>/applications/hive/user/hadoop:
Searching for “endangeredPlantSpecies.q” in the log file will show us it is starting and then beginning the compile phase:
2017-12-22T10:02:00,209 INFO [ff202b74-8efe-46eb-b715-5849a72caa45 main([])]: s3n.S3NativeFileSystem (S3NativeFileSystem.java:open(1210)) - Opening 's3://loggly-emr/scripts/endangeredPlantSpecies.q' for reading 2017-12-22T10:02:00,261 INFO [ff202b74-8efe-46eb-b715-5849a72caa45 main([])]: conf.HiveConf (HiveConf.java:getLogIdVar(3947)) - Using the default value passed in for log id: ff202b74-8efe-46eb-b715-5849a72caa45 2017-12-22T10:02:00,323 INFO [ff202b74-8efe-46eb-b715-5849a72caa45 main([])]: ql.Driver (Driver.java:compile(429)) - Compiling command(queryId=hadoop_20171222100200_95a19046-d8ad-4625-8182-f3a2e9014040): SELECT family, COUNT(species) AS number_of_endangered_species FROM threatened_species WHERE kingdom = 'Plantae' AND "threatened status" = 'Endangered' |
Then, the semantic analysis of the query starting:
2017-12-22T10:02:01,561 INFO [ff202b74-8efe-46eb-b715-5849a72caa45 main([])]: parse.CalcitePlanner (SemanticAnalyzer.java:analyzeInternal(11130)) - Starting Semantic Analysis 2017-12-22T10:02:01,569 INFO [ff202b74-8efe-46eb-b715-5849a72caa45 main([])]: parse.CalcitePlanner (SemanticAnalyzer.java:genResolvedParseTree(11076)) - Completed phase 1 of Semantic Analysis 2017-12-22T10:02:01,569 INFO [ff202b74-8efe-46eb-b715-5849a72caa45 main([])]: parse.CalcitePlanner (SemanticAnalyzer.java:getMetaData(1959)) - Get metadata for source tables 2017-12-22T10:02:01,664 INFO [ff202b74-8efe-46eb-b715-5849a72caa45 main([])]: parse.CalcitePlanner (SemanticAnalyzer.java:getMetaData(2095)) - Get metadata for subqueries 2017-12-22T10:02:01,676 INFO [ff202b74-8efe-46eb-b715-5849a72caa45 main([])]: parse.CalcitePlanner (SemanticAnalyzer.java:getMetaData(2119)) - Get metadata for destination tables 2017-12-22T10:02:01,683 INFO [ff202b74-8efe-46eb-b715-5849a72caa45 main([])]: ql.Context (Context.java:getMRScratchDir(460)) - New scratch dir is hdfs://ip-10-0-8-31.ec2.internal:8020/tmp/hive/hadoop/ff202b74-8efe-46eb-b715-5849a72caa45/hive_2017-12-22_10-02-00_335_6104170905032341771-1 2017-12-22T10:02:01,688 INFO [ff202b74-8efe-46eb-b715-5849a72caa45 main([])]: parse.CalcitePlanner (SemanticAnalyzer.java:genResolvedParseTree(11081)) - Completed getting MetaData in Semantic Analysis |
Finally, it will show a failure, reported by the keyword “ERROR,” followed by a Java error dump:
2017-12-22T10:02:02,800 ERROR [ff202b74-8efe-46eb-b715-5849a72caa45 main([])]: parse.CalcitePlanner (CalcitePlanner.java:genOPTree(423)) - CBO failed, skipping CBO org.apache.hadoop.hive.ql.parse.SemanticException: Line 1:7 Expression not in GROUP BY key 'family' at org.apache.hadoop.hive.ql.parse.SemanticAnalyzer.genAllExprNodeDesc(SemanticAnalyzer.java:11620) ~[hive-exec-2.3.2-amzn-0.jar:2.3.2-amzn-0] … … ... |
And then the actual error message:
(SessionState.java:printError(1126)) - FAILED: SemanticException [Error 10025]: Line 1:7 Expression not in GROUP BY key 'family' |
Now we see the reason for the step failing: there is a field missing from the GROUP BY clause in the SQL statement
Conclusion
Obviously, if we re-submit the job with the correct syntax, it will succeed. But what about other errors in the future? This example was a simple demonstration. In real life, there may be dozens of steps with complex logic, each generating very large log files. Manually sifting through thousands of lines of log may not be practical for troubleshooting purposes. Naturally, the following questions arise:
- Is there any way we can consolidate all the logs in one place?
- Is there any simple way to search for errors?
- Can we be alerted when an error is logged?
- Can we try to find a failure pattern from the logs?
This is where a tool like Loggly can help. We can use it to troubleshoot our EMR job failures. We’ll explore this topic in the third and final part of this blog series.
Ready to give Loggly a try now? Go ahead and sign up for a 14-day trial of the Enterprise tier here and give it a spin.
The Loggly and SolarWinds trademarks, service marks, and logos are the exclusive property of SolarWinds Worldwide, LLC or its affiliates. All other trademarks are the property of their respective owners.

Sadequl Hussain